The Problem
Off-target reactivity is the single largest cause of failed preclinical drug programs, accounting for 62% of all failures. Despite their presumed specificity, emerging data suggest that up to 25% of lead MAbs display off target binding, often against completely unpredicted and unrelated targets. Comprehensive specificity profiling is needed to better predict off-target binding of MAbs and de-risk preclinical candidates prior to IND submission.
The Solution
The Membrane Proteome Array (MPA) was developed to provide detailed MAb specificity analysis against native-conformation epitopes from a library of > 4,500 human membrane proteins. We used the MPA to screen a panel of novel MAbs raised against SLC2A4 (GLUT4), a 12-TM insulin sensitive glucose transporter. MPA analysis demonstrated no SLC2A4 MAb cross-reactivity against related transporters. However, MAb LM052 showed low level reactivity to the unrelated signaling protein Notch1. This was completely unexpected, as SLC2A4 and Notch1 are structurally unrelated (12-TM vs 1-TM) and share less than 6% sequence identity.
The Explanation
Shotgun Mutagenesis Epitope Mapping
High-resolution epitope mapping gave insight into the cross-reactivity of LM052 for Notch1. The epitope for LM052 was mapped at the amino acid level to a loop-constrained LGXXGP sequence on SLC2A4. Despite being unrelated in sequence and structure, the exact same LGXXGP motif is shared on a disulphide-constrained region of Notch1.
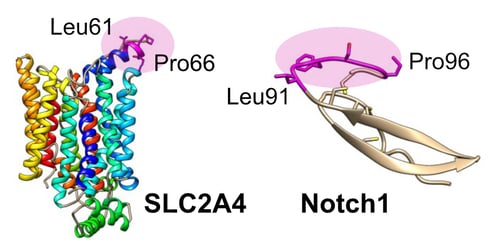
The Impact
Importance of Specificity Testing
The unpredicted cross-reactivity of LM052 with Notch1 attests to the need for comprehensive MAb target profiling and the limitations of specificity testing against family members alone. Our findings indicate that polyspecificity should be assessed prior to the start of preclinical development and selection of lead candidates. (Data featured in Tucker et al, 2018, PNAS)